Getting Started with CMS NanoAOD Open Data
- "What is CMS NanoAOD?"
- "OK! What is in the CMS NanoAOD data?"
- "Nice! But how do I analyse these data?"
- "How can I add quantities to NanoAOD?"
"What is CMS NanoAOD?"
"NanoAOD" is a new data format created by CMS for Run 2 physics analyses. It is available for data collected in 2016 and later. In contrast to the AOD) and MiniAOD formats, NanoAOD is not based on CMS-specific C++ classes, but rather stores data in standard C++ types within a ROOT TTree. You therefore do not need to use the CMS Virtual Machine or docker container to analyze NanoAOD data. NanoAOD can be analyzed using the ROOT program and/or python libraries capable of interpreting the ROOT's TTree structure.
- ROOT: the ROOT program can be installed on your local machine, or you can use the ROOT docker container.
- Python: libraries that are useful for interpreting ROOT file formats are found in the Scikit-HEP project (particularly
uprootandawkward). A python docker container with access to all the required packages is also available.
NanoAOD data can be accessed by:
- Direct download: after identifying an Open Data Portal record, download the files using the cernopendata-client. The "Downloading data files" method will download all the files of a certain record to the local machine.
- XrootD: ROOT contains a network streaming protocol for reading data files called XrootD. A file list can be obtained from the bottom of a dataset's Open Data Portal record (example), or by using
cernopendata-client. Once you know the path to a NanoAOD file, you can access it over the network using ROOT. The following example is done inside the ROOT docker container:
$ root -l root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root
root [0]
Attaching file root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root as _file0...
Warning in <TClass::Init>: no dictionary for class edm::Hash<1> is available
Warning in <TClass::Init>: no dictionary for class edm::ParameterSetBlob is available
Warning in <TClass::Init>: no dictionary for class edm::ProcessHistory is available
Warning in <TClass::Init>: no dictionary for class edm::ProcessConfiguration is available
Warning in <TClass::Init>: no dictionary for class pair<edm::Hash<1>,edm::ParameterSetBlob> is available
(TFile *) 0x561a48eaade0
Within a ROOT script, the TFile::Open method can be used to access the file:
root [2] TFile *f = TFile::Open("root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root")
"OK! What is in the CMS NanoAOD data?"
Each NanoAOD file contains several TTree objects containing different information about the data. This file format is documented in the CMS SWGuide Workbook. The most significant for data analysis is the Events tree, followed by the Runs tree.
$ root -l root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root
root [0]
Attaching file root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root as _file0...
root [1] _file0->ls()
TNetXNGFile** root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root
TNetXNGFile* root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root
KEY: TObjString tag;1 Collectable string class
KEY: TTree Events;1 Events
KEY: TTree LuminosityBlocks;1 LuminosityBlocks
KEY: TTree Runs;1 Runs
KEY: TTree MetaData;1 Job metadata
KEY: TTree ParameterSets;1 Parameter sets
Within each tree, different "branches" represent quantities of interest whose information is stored for each collision event (real or simulated) in the dataset. When NanoAOD files are produced, the branches are all given short documentation strings to aid the user that are collected into a webpage:
Let's see what a NanoAOD file looks like from the ROOT command line, using the docker container. Follow the commands shows above to open a DoubleMuon NanoAOD file using XrootD.
root [6] Events->Show(2) # prints tree contents for event #2.
======> EVENT:2
run = 281616
luminosityBlock = 7
event = 386901
...
...
nMuon = 2
Muon_dxy = -0.00117493,
0.00658798
Muon_dxyErr = 0.0996094,
0.0996094
Muon_dxybs = -0.00116634,
0.00656891
Muon_dz = -0.225708,
0.487549
Muon_dzErr = 4.6875,
5.1875
Muon_eta = 0.902222,
0.339417
...etc...
This shows the basic structure of the NanoAOD file. For event #2, some branches hold a single value for the event (nMuon = 2) while other branches hold an array of values for the event.
In this event the pseudorapidity (eta) values for the 2 muons are stored in an array with 2 elements.
"Nice! But how do I analyse these data?"
The first step to analyzing NanoAOD data is to build up an understanding of what the different branches mean for physics, and how typical CMS physics analyses use these quantities to select interesting events and reconstruct particle decays. Several resources are available to help:
- CMS Open Data Guide: a user's manual for the CMS Open Data with pages covering each of the major physics objects in CMS and many common analysis features such as uncertainties.
- CMS Open Data Workshops: a series of workshops to train researchers to analyze CMS Open Data. Most workshops have included a simplified analysis lesson that uses files in a NanoAOD-like format.
- NanoAOD SWGuide Workbook page: a documentation page for the NanoAOD file format, with an overview of the major physics objects included and links to the production software. Note: this is a wiki site that is updated periodically by CMS and may contain newer information than is relevant for Open Data, and links on the page may point to internal CMS resources.
The second step is to determine a software method for performing validated event filtering, event selection and subsequent analysis. Note: Collision data in NanoAOD format has not been filtered to contain only the validated run segments. A json file containing validated run segments can be found in the links on each collision dataset record.
NanoAOD on the ROOT command line
As we've mentioned, NanoAOD data is stored using ROOT's TTree object. The data can be explored interactively by drawing histograms of various tree branches, even applying weights and cuts to the histograms. In the ROOT container, start the VNC server for X11 graphics support. The default password is cms.cern for either a VNC viewer or the web browser. Then open a NanoAOD file using XRootD and follow these example commands to draw histograms.
$start_vnc
Starting VNC server...
[1] 46
[2] 50
[3] 51
[4] 52
VNC connection points:
VNC viewer address: 127.0.0.1::5901
HTTP access: http://127.0.0.1:6080/vnc.html
$ root -l root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root
root [0]
Attaching file root://eospublic.cern.ch//eos/opendata/cms/Run2016H/DoubleMuon/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/2510000/127C2975-1B1C-A046-AABF-62B77E757A86.root as _file0...
Warning in <TClass::Init>: no dictionary for class edm::Hash<1> is available
Warning in <TClass::Init>: no dictionary for class edm::ParameterSetBlob is available
Warning in <TClass::Init>: no dictionary for class edm::ProcessHistory is available
Warning in <TClass::Init>: no dictionary for class edm::ProcessConfiguration is available
Warning in <TClass::Init>: no dictionary for class pair<edm::Hash<1>,edm::ParameterSetBlob> is available
(TFile *) 0x55f88402e060
root [1] TTree *t = (TTree*)_file0->Get("Events")
(TTree *) 0x55f8837d3910
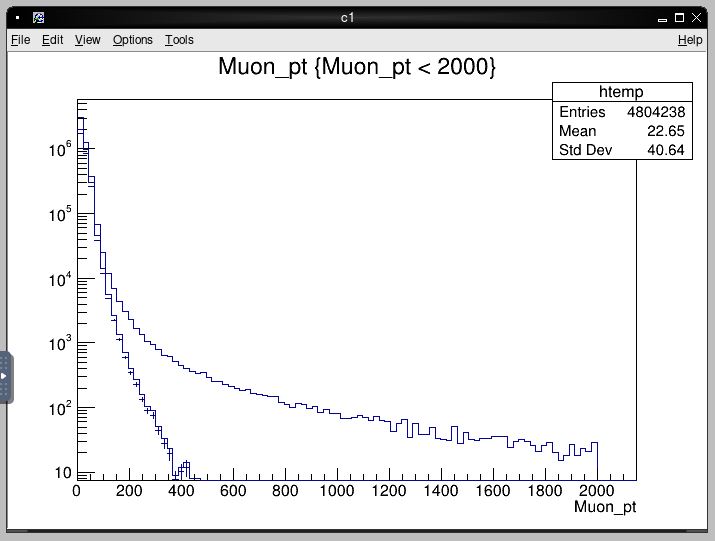
root [2] t->Draw("Muon_pt","Muon_pt < 2000") # draws pT of all muons below 2 TeV
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
(long long) 4804238
root [3] c1->SetLogy(1) # log scale y-axis
root [4] t->Draw("Muon_pt","Muon_tightId > 0","same") # draws pT of high-purity muons
(long long) 3340305
root [5] t->Draw("Muon_pt","(0.85)*(Muon_isTight > 0)","same") # apply an arbitrary weight
(long long) 3340305
root [6] .q
Before exiting the ROOT session, your histogram canvas should look something like this:
NanoAOD skimming and analysis with NanoAOD-tools
The NanoAOD-tools software provides a toolkit to perform operations on NanoAOD files outside the CMSSW software environment. This program can be used to:
- skim events that fail the validation
- skim events that fail an analysis condition based on NanoAOD branch contents
- drop branches from the NanoAOD file to reduce its size
- compute many common corrections for simulation
- compute new observables in user-created modules
Skimming data events that fail the validation is a critical feature, so let's see how to do it by following instructions from the NanoAOD-tools repository. This example will be performed in the ROOT docker container using files from the Tau 2016H dataset. The instructions here download files via git clone, but it is also possible to download a .zip file of the NanoAOD-tools repository from the record page
$ git clone https://github.com/cms-opendata-analyses/nanoAOD-tools.git NanoAODTools
Cloning into 'NanoAODTools'...
remote: Enumerating objects: 4269, done.
remote: Counting objects: 100% (110/110), done.
remote: Compressing objects: 100% (38/38), done.
remote: Total 4269 (delta 57), reused 93 (delta 57), pack-reused 4159
Receiving objects: 100% (4269/4269), 113.08 MiB | 20.57 MiB/s, done.
Resolving deltas: 100% (2230/2230), done.
Updating files: 100% (370/370), done.
$ cd NanoAODTools/
/NanoAODTools$ bash standalone/env_standalone.sh build
Build directory created, please source again standalone/env_standalone.sh without the build argument.
/NanoAODTools$ source standalone/env_standalone.sh
Standalone environment set.
Now we will download the validated runs file that is linked from the Tau dataset record page:
$ wget https://opendata-qa.cern.ch/record/14220/files/Cert_271036-284044_13TeV_Legacy2016_Collisions16_JSON.txt
--2024-01-24 20:01:12-- https://opendata.cern.ch/record/14220/files/Cert_271036-284044_13TeV_Legacy2016_Collisions16_JSON.txt
Resolving opendata-qa.cern.ch (opendata-qa.cern.ch)... 188.185.10.243, 188.185.35.172, 188.185.22.9
Connecting to opendata-qa.cern.ch (opendata-qa.cern.ch)|188.185.10.243|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 11686 (11K) [text/plain]
Saving to: ‘Cert_271036-284044_13TeV_Legacy2016_Collisions16_JSON.txt’
Cert_271036-284044_13TeV_Legacy2016_Collisions 100%[===============================================>] 11.41K --.-KB/s in 0s
2024-01-24 20:01:13 (126 MB/s) - ‘Cert_271036-284044_13TeV_Legacy2016_Collisions16_JSON.txt’ saved [11686/11686]
NanoAOD-tools is based on a "postprocessor" script that can perform different options based on the options provided. To skim the file according to the validated runs json file
we will use the -J option. In the following example, the 2 GB NanoAOD file has been downloaded locally using xrdcp to speed up processing time. To process the file without downloading it, use the full root://eospublic file path shown in the xrdcp command when running nano_postproc.py.
$ xrdcp root://eospublic.cern.ch//eos/opendata/cms/Run2016H/Tau/NANOAOD/UL2016_MiniAODv2_NanoAODv9-v1/260000/03DD0FB3-219B-C54D-A476-EE8937CC214E.root .
[2.156GB/2.156GB][100%][==================================================][11.5MB/s]
$ python scripts/nano_postproc.py -J Cert_271036-284044_13TeV_Legacy2016_Collisions16_JSON.txt ./ 03DD0FB3-219B-C54D-A476-EE8937CC214E.root
Will write selected trees to ./
TClass::Init:0: RuntimeWarning: no dictionary for class edm::Hash<1> is available
TClass::Init:0: RuntimeWarning: no dictionary for class edm::ParameterSetBlob is available
TClass::Init:0: RuntimeWarning: no dictionary for class edm::ProcessHistory is available
TClass::Init:0: RuntimeWarning: no dictionary for class edm::ProcessConfiguration is available
TClass::Init:0: RuntimeWarning: no dictionary for class pair<edm::Hash<1>,edm::ParameterSetBlob> is available
Pre-select 2158176 entries out of 2160343 (99.90%)
Selected 2158176 / 2160343 entries from 03DD0FB3-219B-C54D-A476-EE8937CC214E.root (99.90%)
Done ./03DD0FB3-219B-C54D-A476-EE8937CC214E_Skim.root
Total time 4721.5 sec. to process 2160343 events. Rate = 457.6 Hz.
As before, the TClass warnings from ROOT can be safely ignored. The NanoAOD-tools README page shows more options for this command and discusses their "module" format for applying corrections, performing further event selection, and storing user analysis variables in the file.
More example analysis frameworks
The ROOT Trees within NanoAOD files have been a common HEP data structure for many years, and classic "event loop" methods (primarily written in C++) are an effective and understandable model for analyzing NanoAOD. However, as the overall CMS data size has grown, "columnar" analysis methods have become more and more popular within CMS, and can reduce the computing resources needed for the analysis. The core message is that NanoAOD is extremely flexible!
- Event loop: as a generic example, see the ROOT TTree tutorials. In the Open Data context, the example analyses using "NanoAODRun1" derived data can be replicated for newer NanoAOD files.
- RDataframe: ROOT has developed the RDataFrame package to integrate columnar analysis natively. Several outreach examples using older Open Data in NanoAOD format, such as this Higgs analysis example use RDataFrame in the
skim.cxxscriptto select events and reconstruct a particle. Some example analyses in the NanoAODRun1Examples repository also use RDataFrame, in both C++ and Python. - Coffea/Scikit-HEP tools: significant work has gone into developing HEP software tools within a scientific python ecosystem. The Analysis Grand Challenge from the IRIS-HEP project have created an analysis example that reads NanoAOD files to measure the top quark pair production cross section. The example uses 2015 Open Data that was independently processed as NanoAOD, but the methods can be applied to newer NanoAOD files.
"How can I add quantities to NanoAOD?"
CMS expects that approximately 50% of physics analyses would be able to use only the physics and conditions information stored within NanoAOD files. Other analyses may prefer to analyze the MiniAOD format, which contains more information per event, albeit in the complex EDM class format. See the Getting Started with CMS MiniAOD Data page for more details on analyzing MiniAOD.
One specific collection of information that users may wish to add to NanoAOD is the collection of "Particle Flow Candidates". Some collision data is available in this enriched format, called "NanoAOD-PF". A PFNano producer tool is also available with instructions showing how a MiniAOD file can be processed to produce a NanoAOD-PF ROOT file.
That's it! Hope you enjoyed this introduction. Feel free to play around with the rest of the data and write your own analysis code. Learn more in the CMS Open data guide.